

- General
-
Our lab focuses on four major axes of research, using rewired cell cycle circuits in fission yeast to understand various aspects of the operation, integration and evolution of this essential cellular process and to derive general principles that may be relevant to other eukaryotic networks.
- Background
-
The cell cycle is a highly regulated process, and its control mechanisms have been extensively studied. At the heart of this system are cyclin-dependent protein kinases (CDKs), which associate with various cyclins to sequentially bring about DNA replication (S) and mitosis (M). Activity of CDK complexes is modulated by several inputs, including: 1) control of the respective expression, degradation and subcellular localisation of CDK machinery subunits, 2) specific inhibitors, and 3) changes in phosphorylation state of the catalytic subunit. In fission yeast, cell cycle progression relies on a single CDK, Cdc2, which binds to the cyclins Cig2 and Cdc13 to respectively trigger the G1/S and G2/M transitions, while two additional cyclins, Cig1 and Puc1, play more minor roles in G1. Understanding the fundamental architecture of cell cycle progression has been a challenging endeavour that has led to seminal theoretical and experimental findings, but the complexity of the CDK network has made it difficult to fully appreciate the core principles involved.
To address this, we previously rewired the fission yeast cell cycle control, demonstrating the possibility to replace the cell cycle regulatory network by a simple monomolecular circuit consisting of a fusion between Cdc13 and Cdc2 (Coudreuse and Nurse, Nature 2010). This synthetic system, bypassing much of the known regulation, was sufficient to drive the entire cell cycle with no detectable phenotypes. These findings unraveled the underlying simplicity of core cell cycle control in eukaryotes. Using a chemical genetic strategy, we then showed that cell cycle progression can be artificially organised by simply imposing oscillations between low and high CDK activity at G1/S and G2/M, respectively. In fact, the cell cycle sequence could be remodelled solely by altering CDK activity, which was sufficient to reset G2 cells into G1 without intervening mitosis or to trigger simultaneous entry into S and M from G1 phase. The core eukaryotic cell cycle is therefore built on a basic circuit of two thresholds of qualitatively the same CDK activity and lacks inherent directionality. Cell cycle phases are surprisingly independent, and their temporal sequence is primarily imposed by the frequency and amplitude of the CDK oscillator, revealing the unanticipated modularity of the cell cycle engine.
- Cell cycle architecture and variability
-
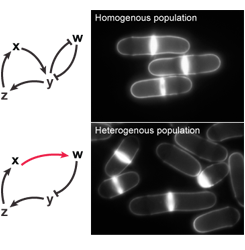
The minimal CDK module (see background section) demonstrates that cell cycle control can be rewired in vivo, providing unexpected insights into the basic architecture of cellular reproduction. Using this system, we investigate the cell cycle engine, focusing on how circuit design quantitatively influences cell cycle progression. Our previous studies suggest that core cell cycle control consists of two layers: 1) qualitative inputs that similarly affect all cells and 2) dedicated regulation or properties of the network that ensure cell-to-cell homogeneity. We are studying this latter control, assessing the reproducibility of cell cycle events between cells by monitoring single-cell and population phenotypes in strains operating with derivatives of the minimal network that have different characteristics. This allows us to isolate critical types of inputs that serve as buffer systems for the variability inherent to the reactions involved in cell cycle control.For these studies, we take advantage of a mathematical model of the minimal cell cycle network developed in collaboration with Dr. Bela Novak (University of Oxford, UK), and use chemical genetics coupled with high resolution imaging by confocal microscopy of live cells grown in microfluidic devices. This work will provide novel insights into the principles of cell cycle control, elucidating the integration of a critical qualitative layer with a quantitative system ensuring cell-to-cell homogeneity.
- Dynamics in cell cycle control
-
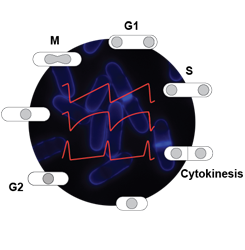 Our understanding of biological events is largely based on static views of molecular interactions, but critical information is often encoded in the dynamic properties of control circuits. In particular, CDK activity dynamics have emerged as a potentially critical input in eukaryotic cell cycle control, in large part through modelling approaches. Hence, bistability and stepwise changes in CDK activity have been proposed to be important components of proper cell cycle progression. However, the complexity of cell cycle regulation, along with the absence of tools to precisely and specifically control CDK activity in vivo with high temporal resolution, has hampered in-depth investigation of this question in living cells.
Our understanding of biological events is largely based on static views of molecular interactions, but critical information is often encoded in the dynamic properties of control circuits. In particular, CDK activity dynamics have emerged as a potentially critical input in eukaryotic cell cycle control, in large part through modelling approaches. Hence, bistability and stepwise changes in CDK activity have been proposed to be important components of proper cell cycle progression. However, the complexity of cell cycle regulation, along with the absence of tools to precisely and specifically control CDK activity in vivo with high temporal resolution, has hampered in-depth investigation of this question in living cells.
To investigate this, we have generated an array of synthetic circuits driving the fission yeast cell cycle with different efficacies as well as constructed new highly tuneable CDK modules that allow us to finely control CDK activity levels. For this project, we utilise specifically-designed microfluidic devices for the control of the cellular environment with a very high temporal resolution, thus combining live-cell imaging with real-time alteration of CDK levels. This will enable us to determine the biological importance of such dynamics and the effects of their alterations on cell cycle progression. - Complexity in cell cycle control
-
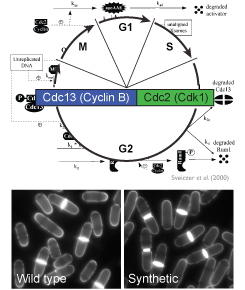
The surprising absence of phenotypes in cells operating with the original synthetic circuit raises the fundamental question of why evolution has selected the complex cell cycle architecture operating in modern cells over simpler and apparently fully functional networks. We are investigating the advantages inherent to the multi-layered control of the cell cycle at two levels. Indeed, simplification of the CDK control circuit may have: 1) long-term effects that alter population growth over several generations and 2) short-term effects, directly impairing functions in specific conditions. This allows us to understand the impact of cell cycle complexity on the fitness of a population of cells and to establish a modular organisation of this network, separating its essential role in cell cycle progression, which solely requires a minimal core system, from more specific functions involving additional inputs.
Our approach relies on in-depth comparison of wild type and synthetic strains operating with cell cycle circuits of different architectures, followed by investigation of the processes that may be turned into limiting inputs by simplification of the circuit. We are also engaged in unbiased approaches to identify unexpected advantages provided by the fission yeast cell cycle circuit as we know it, and the study of the interplay between such functions and cell cycle control represents an important focus of the lab. This project will therefore participate to answering the fundamental question of the rationale behind the evolution of multiple and complex levels of cell cycle regulation. - Evolution of minimal circuits
-
 The complex circuits operating in modern eukaryotic cells often result from alterations and rewiring events that have remodelled simpler networks. Besides species comparisons, the tools to study the evolution of cellular functions toward alternative or more multi-layered regulation remain limited, particularly for essential processes, and the complexity of biological functions has often made difficult the interpretation of controlled evolution experiments.
One of the most fundamental questions in biology concerns the emergence of the “modern” systems that sustain cellular reproduction. We are taking an unprecedented approach to this issue, stepping back through time by using our simple synthetic networks as entry points to understand evolution of the cell cycle. Although these circuits are unlikely to represent the way primitive eukaryotes regulated their division cycle, they provide a theoretical framework for deciphering how this process evolved.
The complex circuits operating in modern eukaryotic cells often result from alterations and rewiring events that have remodelled simpler networks. Besides species comparisons, the tools to study the evolution of cellular functions toward alternative or more multi-layered regulation remain limited, particularly for essential processes, and the complexity of biological functions has often made difficult the interpretation of controlled evolution experiments.
One of the most fundamental questions in biology concerns the emergence of the “modern” systems that sustain cellular reproduction. We are taking an unprecedented approach to this issue, stepping back through time by using our simple synthetic networks as entry points to understand evolution of the cell cycle. Although these circuits are unlikely to represent the way primitive eukaryotes regulated their division cycle, they provide a theoretical framework for deciphering how this process evolved.
This project is based on laboratory evolution experiments in which various synthetic fission yeast strains with different cell cycle properties are grown in selected environments. This takes advantage of multiplexed systems that allow us to maintain these cultures under controlled conditions (implemented with the kind support of Dr. Maitreya Dunham, Washington University, Seattle, USA). Emerging clones with improved proliferation potential compared to the starting cells are then isolated in different conditions, and we will study how cell cycle improvement is brought about.This approach also allows us to extract more general principles in the evolution of essential cellular functions.

